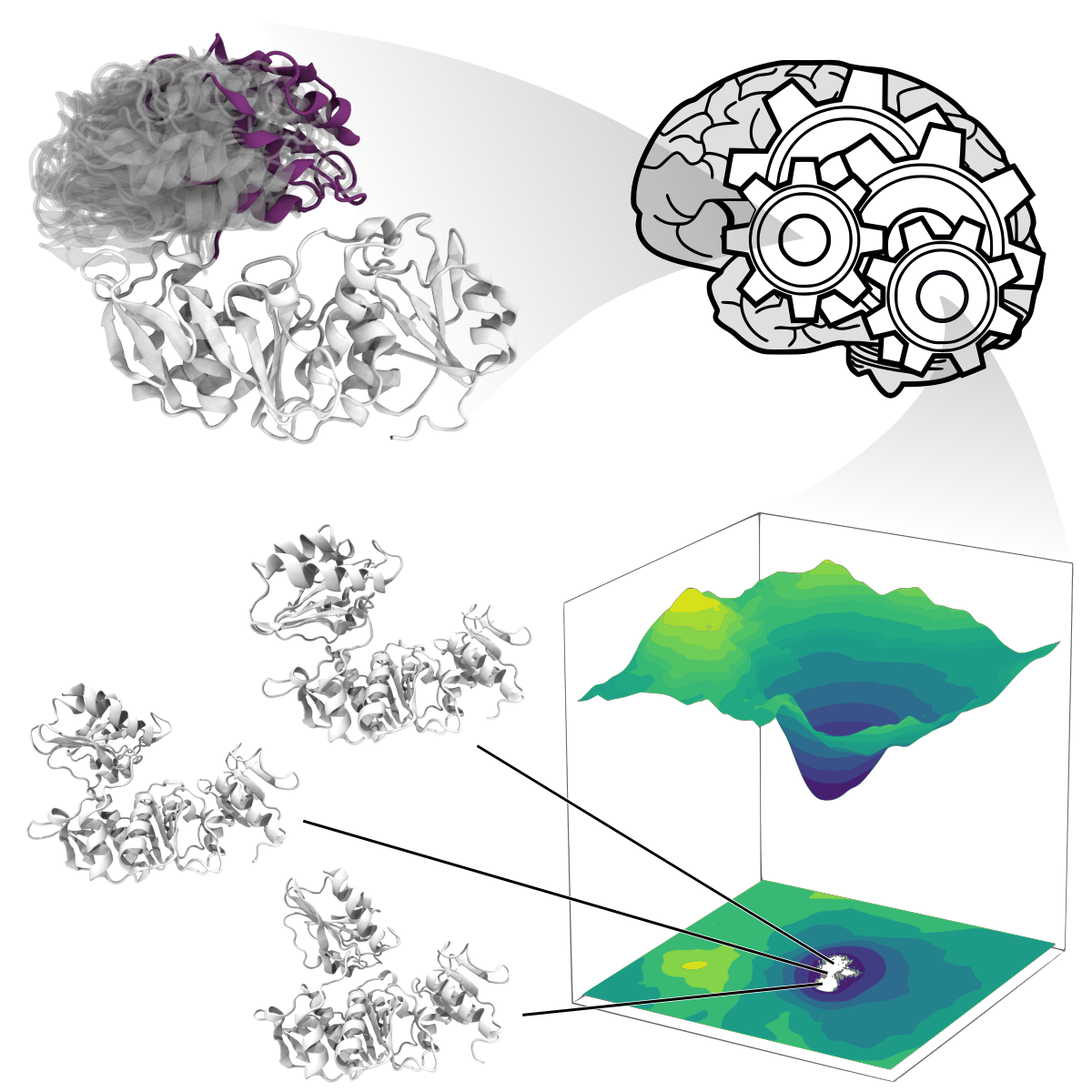
 Structural variability plays a central role in biology. Conformational dynamics enable proteins to react to changes in their environment and to interact with designated binding partners, while evolutionary variability enables proteins to develop new functionalities. We create and apply methodologies to characterise and predict atomic-level protein structural variability. In this context, we are showing that generative neural networks can be leveraged to characterise protein conformational space, enabling the prediction of possible transition states and identification of suitable conformations for protein docking.
Structural variability plays a central role in biology. Conformational dynamics enable proteins to react to changes in their environment and to interact with designated binding partners, while evolutionary variability enables proteins to develop new functionalities. We create and apply methodologies to characterise and predict atomic-level protein structural variability. In this context, we are showing that generative neural networks can be leveraged to characterise protein conformational space, enabling the prediction of possible transition states and identification of suitable conformations for protein docking.
Representative articles:
- MS.C. Musson and M.T. Degiacomi (2023), Molearn: a Python package streamlining the design of generative models of biomolecular dynamics, Journal of Open Source Software 8(89) 5523
- V.K. Ramaswamy, S.C. Musson, C.G. Willcocks, M.T. Degiacomi (2021). Learning protein conformational space with convolutions and latent interpolations, Physical Review X
- M.T. Degiacomi (2019), Coupling Molecular Dynamics and Deep Learning to Mine Protein Conformational Space, Structure