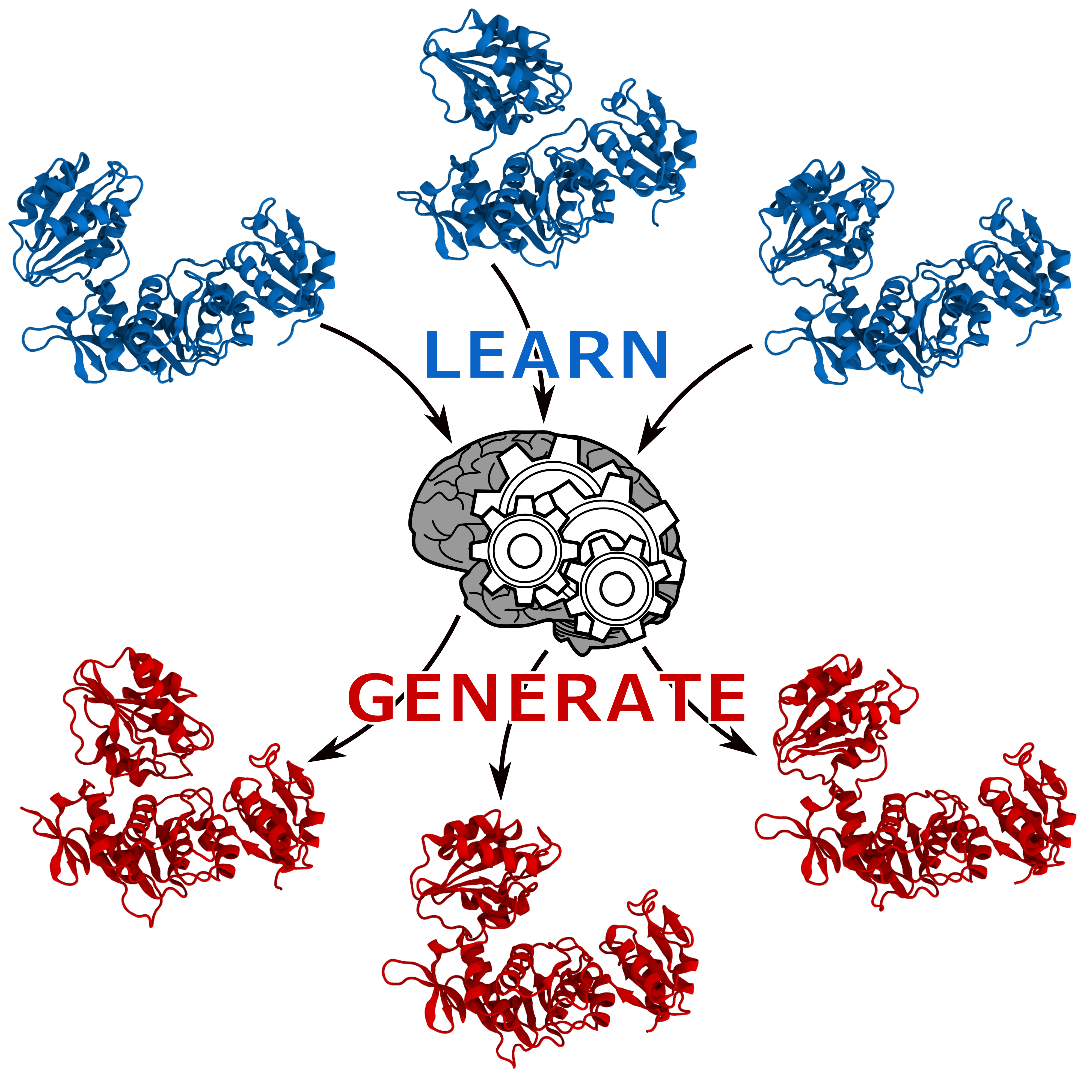
 In recent years, generative neural networks have been gaining popularity owing to their ability to produce believable fake data including photos, video and even news. The creativity of these neural networks lies on their capacity of generating something new based on collection of examples provided as input. We show that autoencoders, a kind of neural network, can be exploited to generate meaningful protein conformations.
In recent years, generative neural networks have been gaining popularity owing to their ability to produce believable fake data including photos, video and even news. The creativity of these neural networks lies on their capacity of generating something new based on collection of examples provided as input. We show that autoencoders, a kind of neural network, can be exploited to generate meaningful protein conformations.
Protein molecular function and malfunction in an organism is often linked to their interconversion between states caused by changes in environmental conditions or binding of ligands such as drugs or other proteins. In this context, we demonstrate two possible usages for an autoencoder: (1) predicting the transition path between two states sampled by MD simulations, when no sampling of the intermediates is available; (2) coupling the autoencoder with POWer, our protein docking algorithm, to help the prediction of proteins’ arrangement into a complex when the subunits undergo substantial conformational change.