Eager to learn about how mass spectrometry can help structural biology? Join us in Bordeaux on the 5-7 October 2020 to learn everything about top-down and native MS, ion mobility, HDX, cross-linking and, of course, integrative modelling!
Blog
Welcome Holly and George!
This week two Masters students joined our group. George, from Chemistry department, will work on expanding JabberDock capabilities while Holly, from Physics, will study protein conformational spaces via deep learning. Welcome both!
Moving to Physics
Today the group moves to Durham University Department of Physics, where Matteo will take the position of Assistant Professor in Condensed Matter Physics!
Protein docking with JabberDock
We are very excited to present JabberDock, our new protein-protein docking algorithm. JabberDock is capable of accommodating for rearrangements upon binding including side chain reorientations and backbone flexibility. To do this, it leverages Spatial and Temporal Intensity (STID) maps, our single volumetric representation for proteins surface, electrostatics and local dynamics. JabberDock is freely available on Github, and is presented in the following article:

This publication not only complies to the Palatinate Challenge but, more importantly, it is the first article of Lucas Rudden, PhD student in our group. Congratulations!
Welcome Lorenza
Today we welcome Lorenza Pacini, PhD student in ENS Lyon under the supervision of Dr. Claire Lesieur and Prof. Laurent Vuillon (Université Savoie Mont Blanc). Lorenza will spend one month with us, developing methods to model protein fibrils. Looking forward to some collaborative software development!
Well done Sam!
Sam successfully completed his Masters project with us, presenting a great poster on his study of peptide-lipid interactions by molecular dynamics simulations. Congratulations!
Chaperone-regulated mechanosensation
Small Heat Shock proteins such as HspB1 are molecular chaperones in charge of preventing harmful misfolding of proteins under stress conditions. Work led by the Benesch and Gehmlich groups shows, from muscle fibers down to single molecules, that phosphorylation of HspB1 alters its intramolecular dynamics, facilitating its binding to the mechanosensitive Filamin C. In good correspondence with NMR data, our calculations reveal that over over the course of microsecond-long simulations the N-terminus of HspB1 detaches from the rest of the protein.
Collier M.T., Alderson, T.R., de Villiers, C.P., Nicholls, D. , Gastall, H.Y., Allison, T.M., Degiacomi M.T., Jiang, H., Mlynek, G., Fürst, D.O., van der Ven, P.F.M., Djinovic-Carugo, K., Baldwin, A.J., Watkins, H., Gehmlich, K., Benesch, J.L.P. (2019), HspB1 phosphorylation regulates its intramolecular dynamics and mechanosensitive molecular chaperone interaction with filamin C. Science Advances, 5(5)
Cover article on JBC!
 Our article was Editors’ Pick Highlight, and image selected as a cover article for the 10th May edition of JBC!
Our article was Editors’ Pick Highlight, and image selected as a cover article for the 10th May edition of JBC!
The image shows the structure of the long-lived small heat-shock proteins (present in the eye lens), and the effect of age-related isomerisation of key amino-acids, as detailed in our article.
[image composed with Gimp from protein VMD rendering redesigned by the artistic hand of Valentina Erastova]
What can one do with a neural network trained on molecular dynamics simulations?
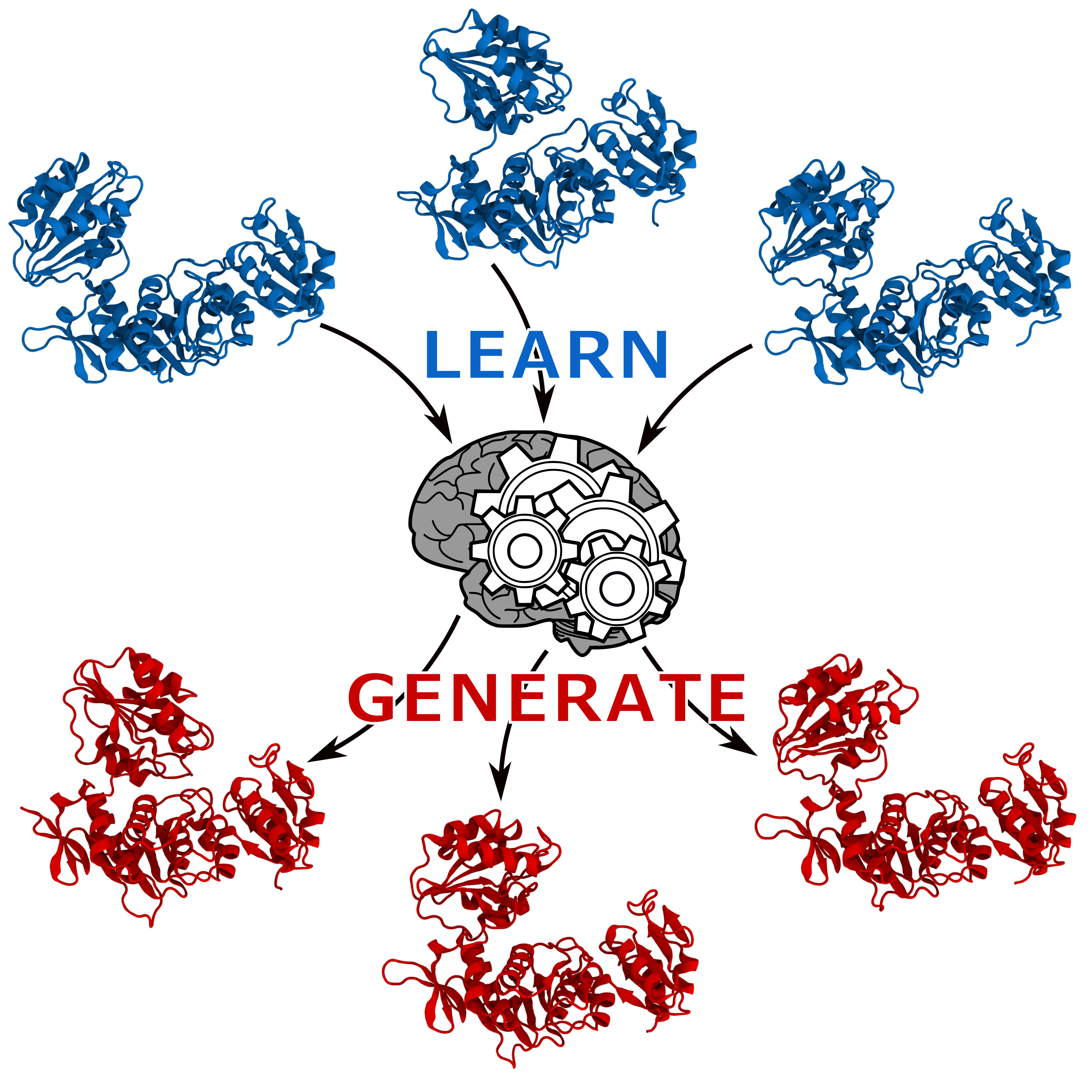 In recent years, generative neural networks have been gaining popularity owing to their ability to produce believable fake data including photos, video and even news. The creativity of these neural networks lies on their capacity of generating something new based on collection of examples provided as input. We show that autoencoders, a kind of neural network, can be exploited to generate meaningful protein conformations.
In recent years, generative neural networks have been gaining popularity owing to their ability to produce believable fake data including photos, video and even news. The creativity of these neural networks lies on their capacity of generating something new based on collection of examples provided as input. We show that autoencoders, a kind of neural network, can be exploited to generate meaningful protein conformations.
Protein molecular function and malfunction in an organism is often linked to their interconversion between states caused by changes in environmental conditions or binding of ligands such as drugs or other proteins. In this context, we demonstrate two possible usages for an autoencoder: (1) predicting the transition path between two states sampled by MD simulations, when no sampling of the intermediates is available; (2) coupling the autoencoder with POWer, our protein docking algorithm, to help the prediction of proteins’ arrangement into a complex when the subunits undergo substantial conformational change.
A minor alteration with profound consequences
Long-lived proteins may accumulate a range of modifications over time, including subtle alterations such as side-chain isomerization.
In collaboration w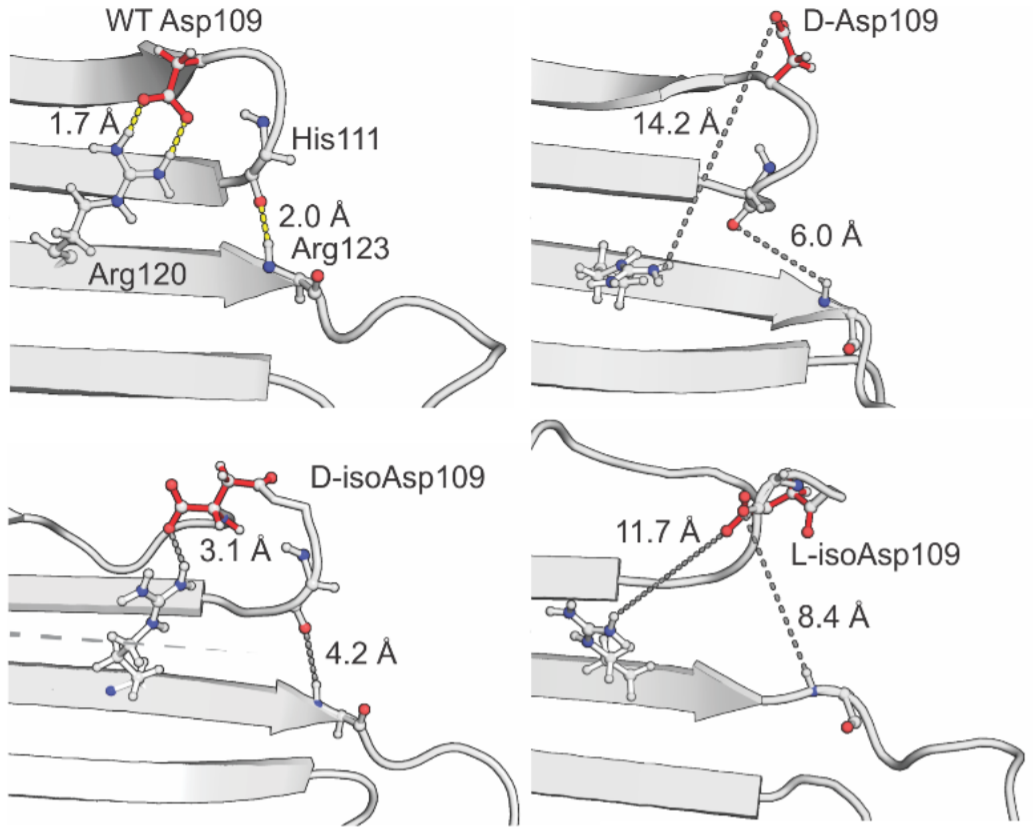 ith the
ith the
Julian group in UC Riverside and Benesch group in Oxford we study the effect of isomerisation of an aspartate residue in αB-crystallin, the most abundant chaperone proteins in the eye lens and within the longest-lived proteins in the body. Malfunction of these proteins is linked to a range of diseases, including cataract.
Our results illustrate how age-related isomerization of amino acid residues, which may seem to be only a minor structural perturbation, can disrupt native structural interactions with profound consequences for protein assembly and activity.
Lyon, Y.A., Collier, M.P., Riggs, D.L., Degiacomi, M.T., Benesch, J.L.P., Julian, R.R. (2019). Structural and functional consequences of age-related isomerization in α-crystallins, Journal of Biological Chemisty